
Table of contents
Met4DX Documentation
Introduction
Met4DX is a fast, robust, and user-friendly mass spectrometry data processing tool for metabolomics and lipidomics. The versatile tool facilitates the processing of both 3-dimensional LC-MS data and 4-dimensional LC-IM-MS data, encompassing main functions such as data conversion, peak detection, retention time correction, peak grouping, assignment of MS/MS spectra, and others.
Specifically, for peak detection, Met4DX employs a unique mass spectrum-oriented bottom-up assembly algorithm, ensuring the high-sensitive and high-coverage detection of TRUE MS peaks in metabolomics data (both 3D and 4D datasets). Additionally, Met4DX also supports a uniform peak quantification strategy that enhances the accuracy of peak quantitation in large-scale metabolomics analysis.
Moreover, Met4DX allows users to export results, enabling further analysis through complementary tools, such as MetDNA for metabolite identification, MetFlow for large-scale data integration, and MetTracer for stable-isotope tracing metabolomics data analysis.
Dependencies
Met4DX depends on the following software or SDKs to read the vendor data. Users who uses Met4DX are considered to agree with the licenses for the listed tools.
Recommended configuration
Data format available for analysis
|
Vendor |
Formats[1] |
Supported |
Tested models |
Acquisition methods |
|
Agilent |
.d/ |
Yes |
Agilent 6550 |
MS1 only |
|
Bruker |
.d/ |
Yes |
timsPro |
PASEF-DDA |
|
Sciex |
.wiff, .wiff2 |
Yes |
Sciex 6600 |
DDA/SegmentDDA |
|
Shimadzu |
.lcd |
Yes |
LCMS-9050 |
DDA |
|
Thermo |
.raw |
Yes |
Exploris 480, Astral, Excedion Pro[2] |
DDA/SegmentDDA |
|
Waters |
.raw/ |
Yes |
Synapt XS |
DDA |
|
.uep |
Yes[3] |
Xevo MRT |
DDA |
[1] A format ending with a slash (‘/’) indicates the data is stored in a folder. If the format does not end with a slash, it represents a standalone file.
[2] It is required to convert the raw data to mzML format using Compound Discoverer software for Orbitrap Excedion Pro instruments before adding to Met4DX is required.
Installation
Met4DX installer supports Windows 10 and above only currently. Please download the installer from the download page and install it directly. Please follow the installation wizard to install the software as you like.
Please contact us via metdna (AT) sioc.ac.cn if you have any problem.
NOTE:
Interface
Initial interface
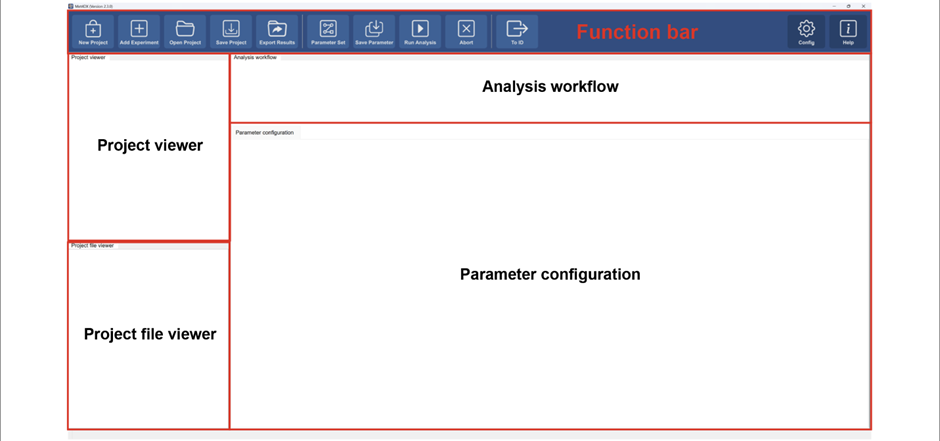
Function bar

Project and data managements
Data processing tools
Open in identification tools
Other functions
Project setting
A project is required for data processing with Met4DX. Project means a bunch of experiments of different LC columns or ionization modes for certain samples acquired using the same MS instrument. Even if you have only one experiment, a project is still required. One should put all data together for the same project and submit the root directory to create the project and add corresponding experiments as follows:
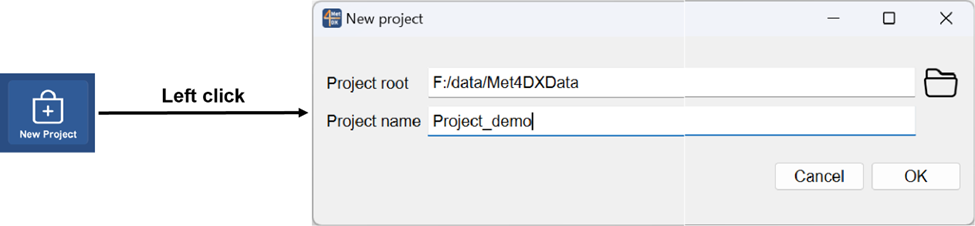
Create a new project
Select the root director of the project root and name the project. If the project name is set as empty, a default project name will be set to the folder name of the project root.
Save current project
Save current project to a file (in .4dx format for Met4DX).

Open a previous project
Open a Met4DX project file (in .4dx format). Besides, Met4DX project file is registered to Met4DX software. Hence, one can double click the saved Met4DX project file to open it with Met4DX software.

Experiment setup
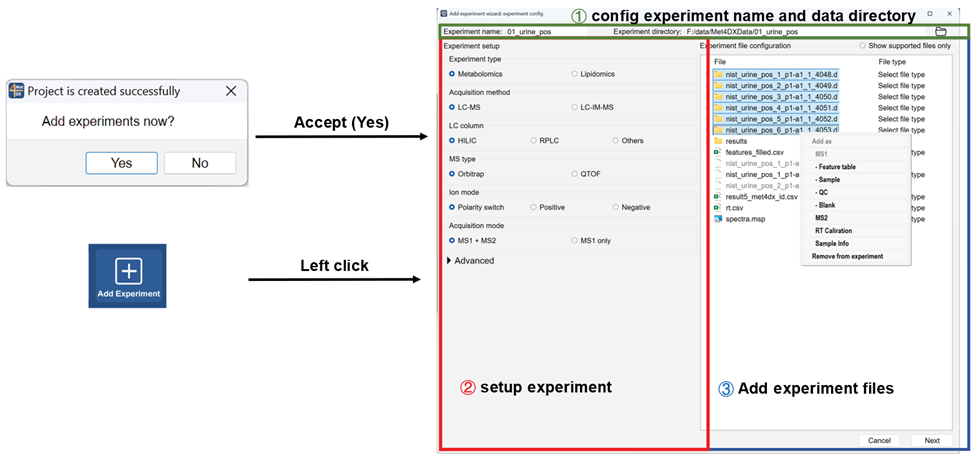
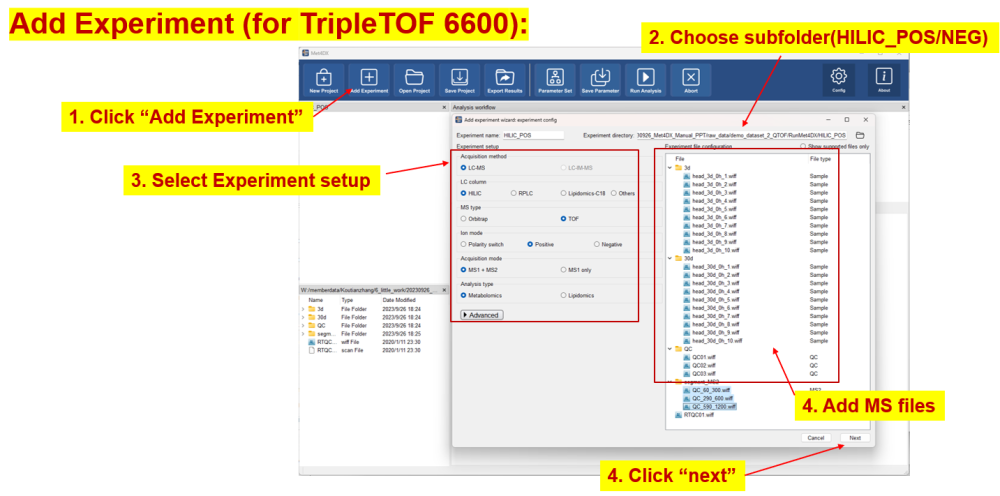
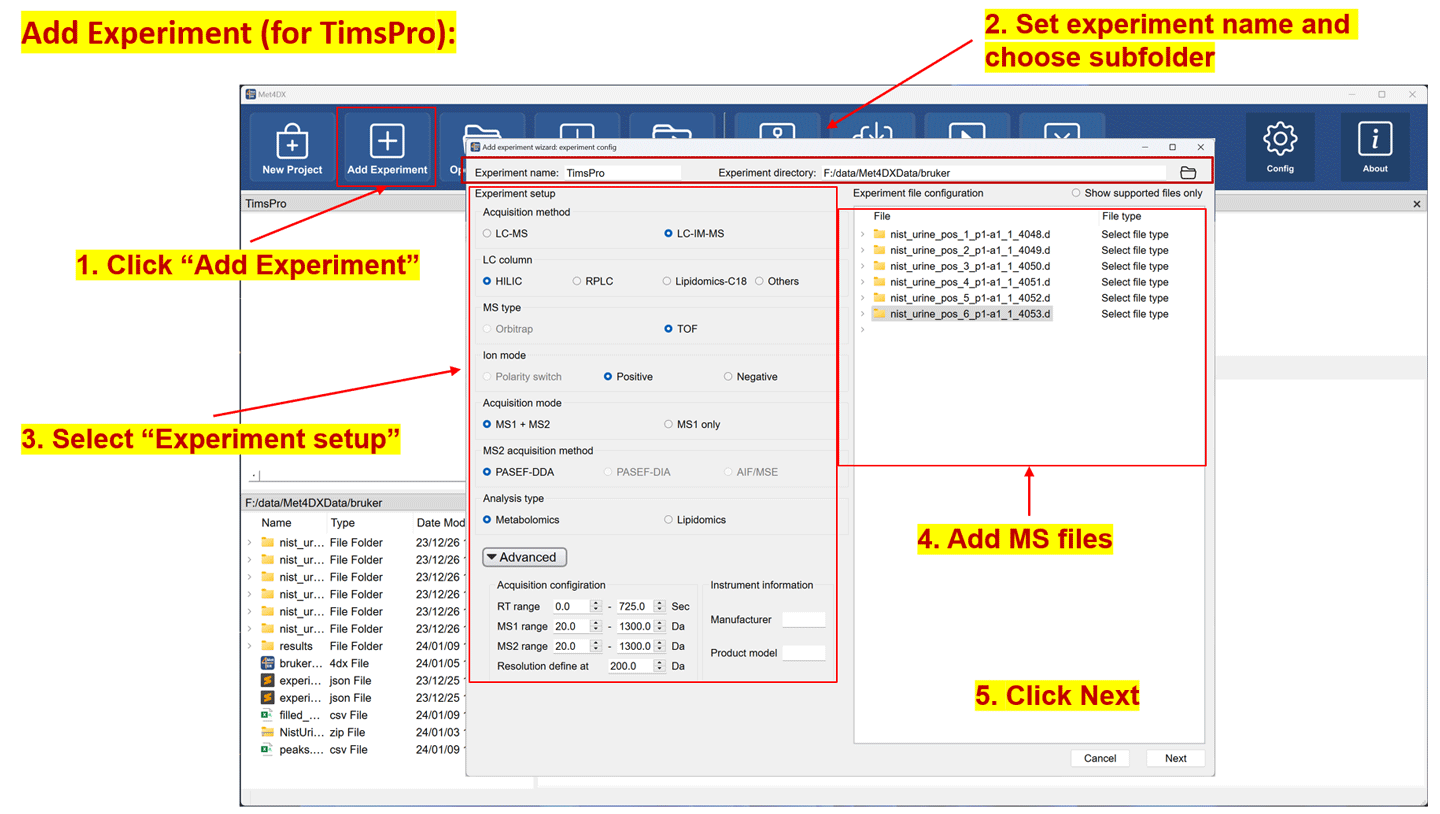
Add experiment
Experiments can be added with the ‘add experiment wizard’ by accepting ‘Add experiments now’ dialogue after creating the project or click the ‘Add experiment’ button from the function bar.
NOTES:
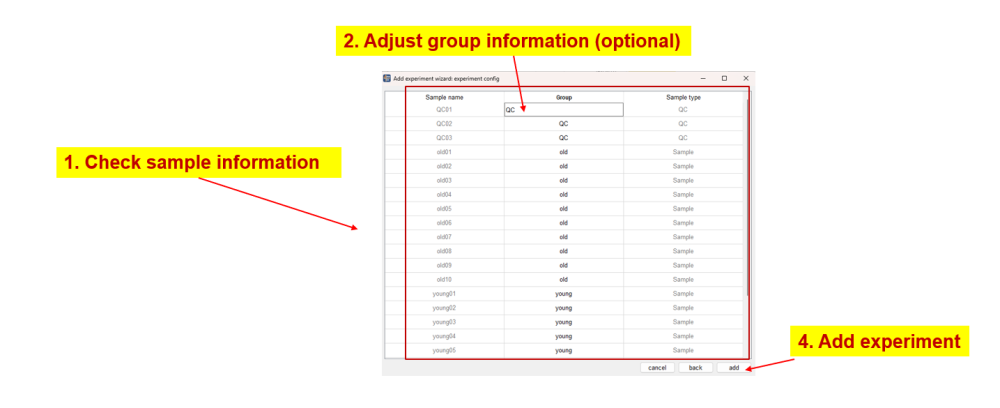
Confirm sample information
The software will check the added files and group samples automatically according to the subfolders of MS1 files in submitted experiments after clicking ‘next’ button on the first page of ‘add experiment wizard’ and show the sample info in the second page. If there is no problem, one can click the ‘add’ button to add the configured experiment. The added experiments will be shown in project viewer in the top left panel of Met4DX.
If you have multiple experiments, please add them sequentially with the wizard.

NOTE:
Parameter setting
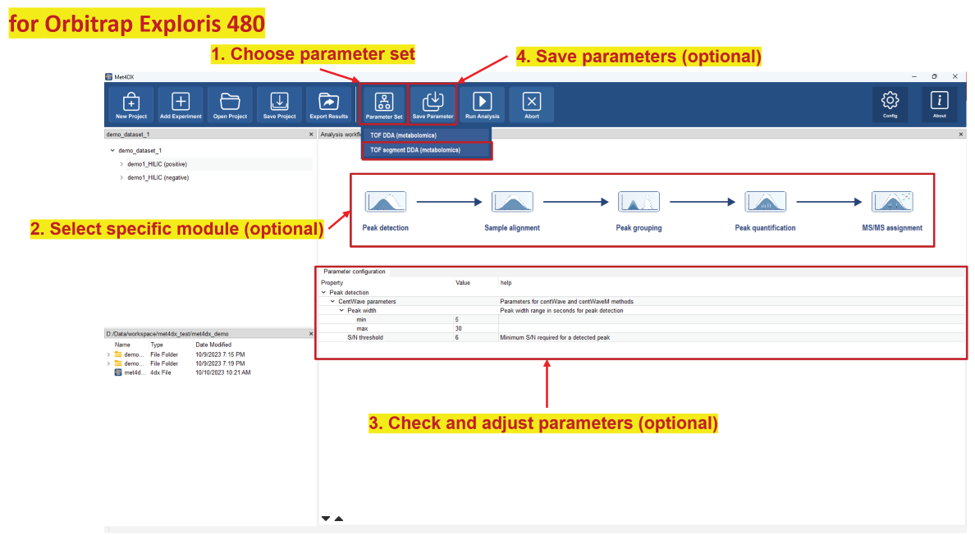
Met4DX provides the most used parameter sets in Zhulab for different data processing tasks, which is highly recommended for inexperienced users, and even for all users. To start a data processing workflow, one should load a parameter set first, modifying parameters later, and submit experiment for data processing at last.
Load a parameter set
A parameter set contains multiple steps for data processing, which will be shown in the analysis workflow viewer after loading a parameter set. Parameters of each step can be configured by clicking corresponding steps. Key parameters will show by default and extended parameters will be shown and hidden by clicking the down and up triangle tool button in the button of the parameter configuration panel, respectively.

Save current parameter set
Users can save customized parameter sets according to actual needs for data processing next time with the data acquired with the same methods. Please configure the parameter set information using the pop-up dialogue window.

Parameter sets
|
LC-MS |
TOF |
MS1 only |
Metabolomics |
TOF MS1 only (metabolomics) |
|
|
Lipidomics |
TOF MS1 only (lipidomics) |
||||
|
MS1+MS2 |
Metabolomics |
DDA[1] |
TOF DDA (metabolomics) |
||
|
Segment DDA[2] |
TOF segment DDA (metabolomics) |
||||
|
Processed[3] |
TOF search MS/MS (metabolomics) |
||||
|
Match MS/MS (metabolomics) |
|||||
|
Lipidomics |
DDA |
TOF DDA (Lipidomics) |
|||
|
Segment DDA |
TOF segment DDA (Lipidomics) |
||||
|
Processed |
TOF search MS/MS (Lipidomics) |
||||
|
Match MS/MS (Lipidomics) |
|||||
|
Orbitrap |
MS1 only |
Metabolomics |
Orbitrap MS1 only (metabolomics) |
||
|
Lipidomics |
Orbitrap MS1 only (lipidomics) |
||||
|
MS1+MS2 |
Metabolomics |
DDA |
Orbitrap DDA (metabolomics) |
||
|
Segment DDA |
Orbitrap segment DDA (metabolomics) |
||||
|
Processed |
Orbitrap search MS/MS (metabolomics) |
||||
|
Match MS/MS (metabolomics) |
|||||
|
Lipidomics |
DDA |
Orbitrap DDA (Lipidomics) |
|||
|
Segment DDA |
Orbitrap segment DDA (Lipidomics) |
||||
|
Processed |
Orbitrap search MS/MS (Lipidomics) |
||||
|
Match MS/MS (Lipidomics) |
|||||
|
LC-IM-MS |
TOF |
MS1 only |
Metabolomics |
IMS MS1 only (metabolomics) |
|
|
Lipidomics |
IMS MS1 only (lipidomics) |
||||
|
MS1+MS2 |
Metabolomics |
Pasef DDA |
IMS DDA MS2 (metabolomics) |
||
|
IMS DDA MS1 (metabolomics) |
|||||
|
Processed |
IMS search MS/MS (metabolomics) |
||||
|
Match MS/MS (metabolomics) |
|||||
|
Lipidomics |
Pasef DDA |
IMS DDA MS2 (lipidomics) |
|||
|
IMS DDA MS1 (lipidomics) |
|||||
|
Processed |
IMS search MS/MS (lipidomics) |
||||
|
Match MS/MS (lipidomics) |
[1] DDA parameter set requires the data to be acquired using the DDA method, in which both MS1 and DDA MS2 are contained, and no additional MS2 files
[2] Segment DDA parameter set requires the data to contain extra MS2 data which is collected using segmented DDA method. In this case, segment DDA data (MS2 files) is submitted as MS2 file.
[3] Processed parameter set are used for experiments which are already processed with the feature table and spectrum data file(s)
NOTES:
Parameter configuration
Parameters for each step are viewed and edited by clicking icon button of the corresponding step. Here is an example view:





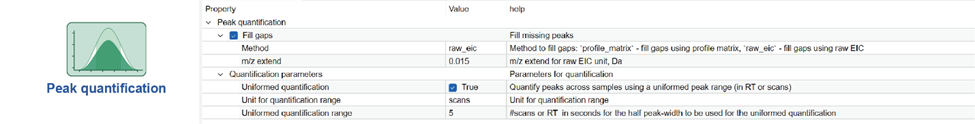
NOTE:
Data processing
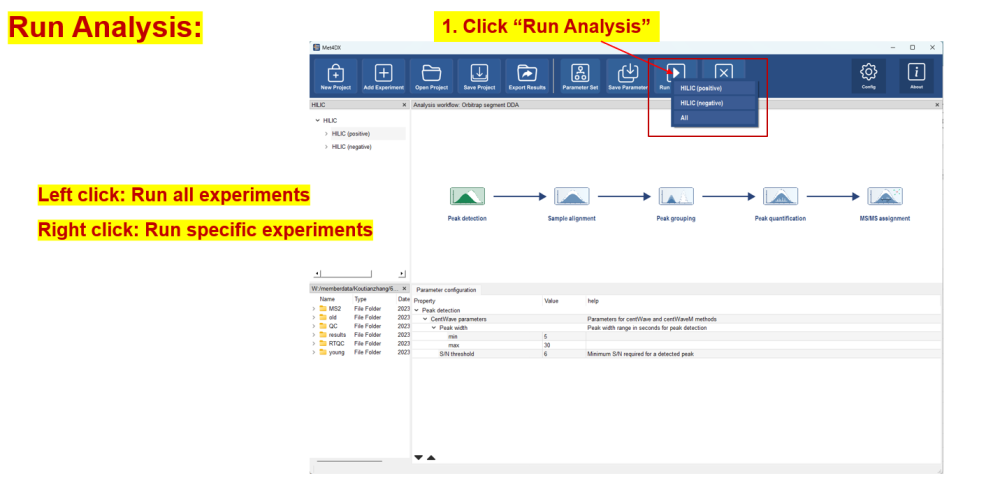
Run analysis
After submission, the experiments in project viewer will show the corresponding processing status.
 current processing experiment.
current processing experiment. submitted experiment, waiting for processing.
submitted experiment, waiting for processing. already processed experiment.
already processed experiment.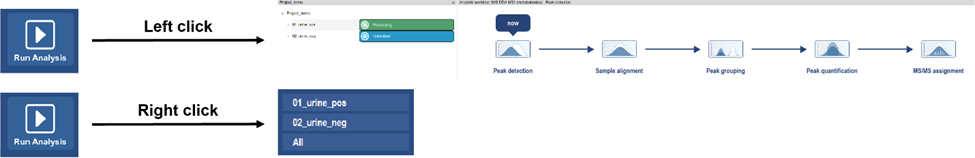
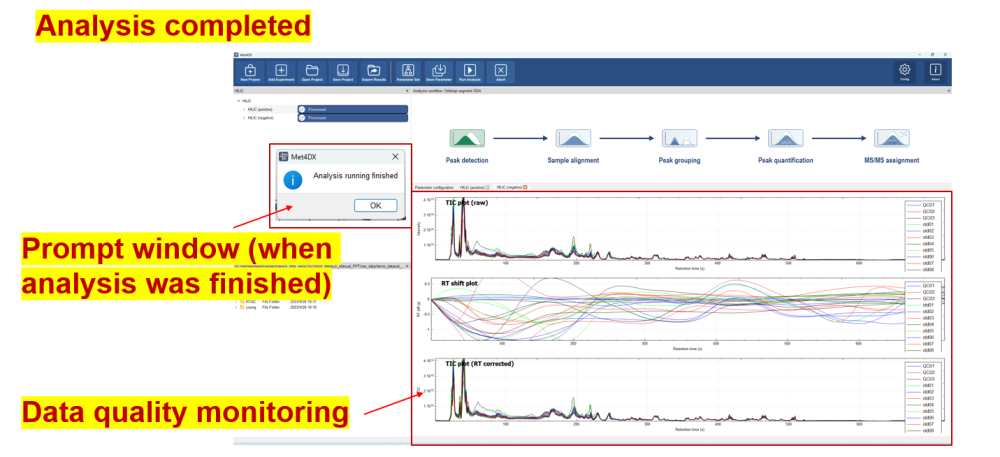
Processing plots
TIC plot (raw), RT shift plot, TIC plot (RT corrected) will be displayed during processing after corresponding steps finished.
These intermediate diagrams can be used to monitor data quality (such as RT shift, TIC consistency, etc.) and whether it has problems in data processing with submitted parameters.

Abort analysis
If you want to stop the analysis process, please click the 'Abort' button. In this case, the current processing task will be stopped and all other submitted but not finished tasks will be aborted.

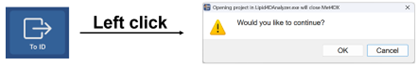
Open in identification tools
For lipidomics analysis, Met4DX now supports lipid identification in Lipid4DAnalyzer tool after raw data processed with MSMS assignment. For lipids identification with Lipid4DAnalyzer, please refer to the documentation of Lipid4DAnalyzer (pdf).
Other functions
View helps documents, check logs and report bugs
Met4DX supports reading the documentation, reporting bugs and show logs and software information through the ‘Help’ menu.
Met4DX results
Default outputs
The data processing results are stored in the results folder under the project folder for the corresponding experiment.

Export results
One can also export results for further analysis with other software tools from Zhulab, such as MetDNA, MetFlow, and even MetTracer for isotope tracing experiments. To export results, please click ‘Export Result’ button and select export type for your need.

NOTE:
Tutorial
Demo datasets
Steps
Demo dataset 1: mouse liver tissues from Orbitrap Exploris 480

Demo dataset 2: fly head tissues from TripleTOF 6600

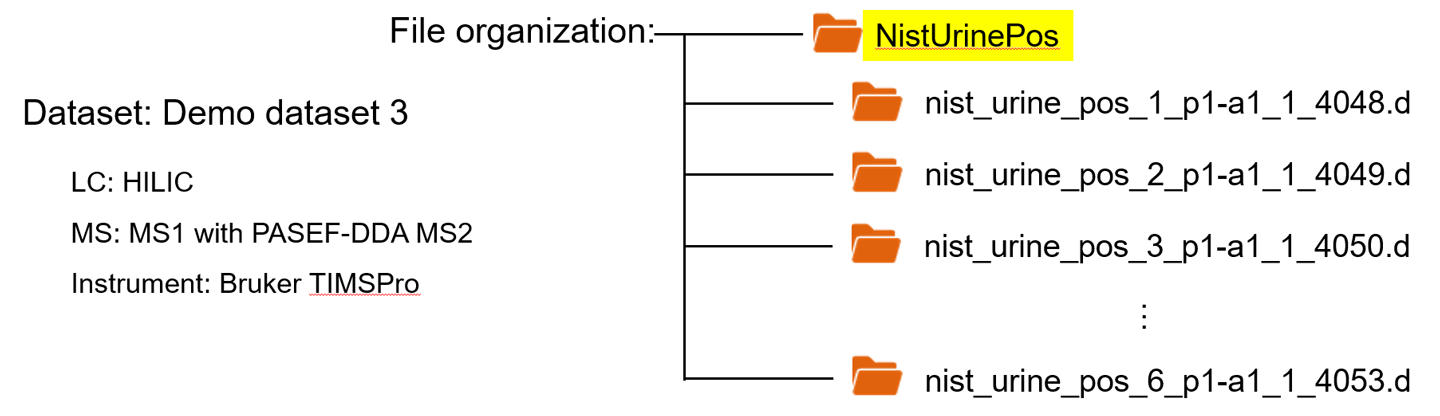
Demo dataset 3: NIST urine positive data for Bruker TIMSPro
Demo dataset 1: mouse liver tissues from Orbitrap Exploris 480

Demo dataset 2: fly head tissues from TripleTOF 6600
Demo dataset 3: NIST urine positive data for Bruker TIMSPro
Demo dataset 1: mouse liver tissues from Orbitrap Exploris 480
Demo dataset 2: fly head tissues from TripleTOF 6600

Demo dataset 3: NIST urine positive data for Bruker TIMSPro




Config Met4DX software

Software configuration
Parameter set config
One can rename, delete, or modify information of the parameter sets saved by the user. Detailed value of parameters can also be configured in this configuration. Please save the parameters before you leave it. Otherwise, the modification will be ignored.
The help column of the parameter configuration view shows the meaning of each parameter. If you have problems, please contact us for clarification.